We study spindle assembly in oocytes and embryos – the dynamics of microtubules, the cooperation of signaling molecules, and the role of newly described pathways.
Contact: David Drutovič
Mammalian oocytes and early embryos are prone to chromosome segregation errors, resulting in unequal distribution of genetic information into daughter cells, which can lead to infertility and developmental defects. The formation of euploid cells, i.e., cells with the correct number of chromosomes, critically depends on the assembly of the meiotic spindle—a cellular structure that ensures proper attachment of chromosomes and their segregation toward the poles of the cell. In recent years, numerous studies showed that spindle defects in human oocytes are a major age-independent factor of increased aneuploidy. Examples of such spindle defects include the inability to maintain a stable spindle and the transient formation of so-called multipolar spindles, which promote incorrect chromosome-microtubule attachments. Given the rising prevalence of infertility, understanding the causes of meiotic spindle defects has clear clinical relevance.
The mechanisms controlling correct spindle assembly in human oocytes and embryos are not yet fully described. In collaboration with other laboratories, we have previously characterized essential factors involved in spindle assembly regulation. We described the key role of the three Aurora kinases, proteins that control both mitotic division in somatic cells and meiotic division in sperm and oocytes. We found that human, as well as mouse chromosomes in oocytes, similarly activate proteins that govern their own division.
This research project aims to elucidate the molecular basis of functional meiotic spindle assembly, ensuring accurate chromosome segregation in oocytes and early embryos. We will use oocytes and embryos from model organisms, primarily mice, to define generally applicable principles in mammals, including humans. To understand spindle assembly, it is essential to investigate the specific roles of signaling proteins and their interactions. We hypothesize that interactions between signaling pathways are crucial for proper spindle formation and chromosome segregation.
To better examine spindle formation in oocytes, we have optimized a three-color time-lapse fluorescence imaging. Light-sheet microscopy allows us to monitor chromosome segregation, spindle formation, and spindle pole dynamics throughout the entire live oocyte and embryo at high resolution. This approach enables us to characterize spindle assembly after inhibition of various signaling pathways using specific pharmacological inhibitors in oocytes and embryos. Time-lapse imaging at various stages of embryonic development enables us to describe the molecular mechanisms that control spindle assembly during early developmental stages.
We will employ state-of-the-art technologies and innovative approaches, including live-cell microscopy, FRET biosensors for monitoring signaling protein activity in living cells, and advanced image analysis using artificial intelligence and machine learning. This will allow us to uncover how individual pathways regulate each other and determine whether they are essential for meiotic progression in oocytes and embryos across different model organisms. We will systematically study less-characterized roles of signaling proteins, investigate changes in spindle assembly in detail, and uncover their function in accurate chromosome segregation in oocytes and embryos.
Our primary goal is to focus on the main causes of chromosome segregation errors during early embryonic development. The results will be necessary for obtaining high-quality oocytes and improving embryo selection for transfer in clinical settings. The knowledge gained could help to better understand developmental abnormalities, improve infertility diagnostics, and develop therapeutic strategies to reduce spindle defects in female meiosis.

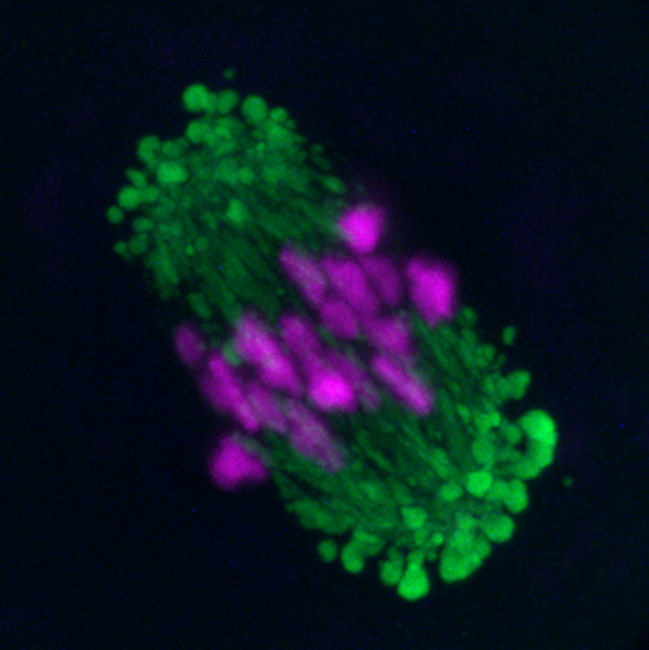
Figure 1: Fluorescence image of a mouse oocyte showing alignment of chromosomes before segregation (magenta) and newly discovered domains around the spindle (green), which support spindle formation
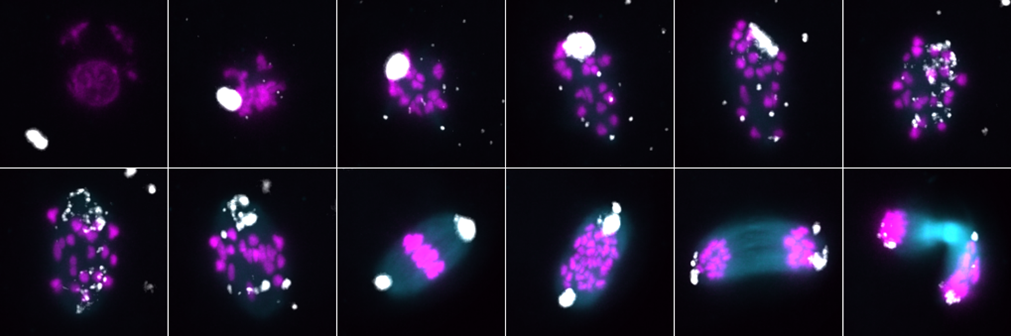
Figure 2. Chromosome segregation (magenta) during mouse oocyte meiosis is mediated by the spindle formation (green). Its assembly depends on so-called MTOC centers (white), which fragment and later cluster before chromosome segregation. The figure shows the time points of oocyte meiosis from its onset to the division of the oocyte.
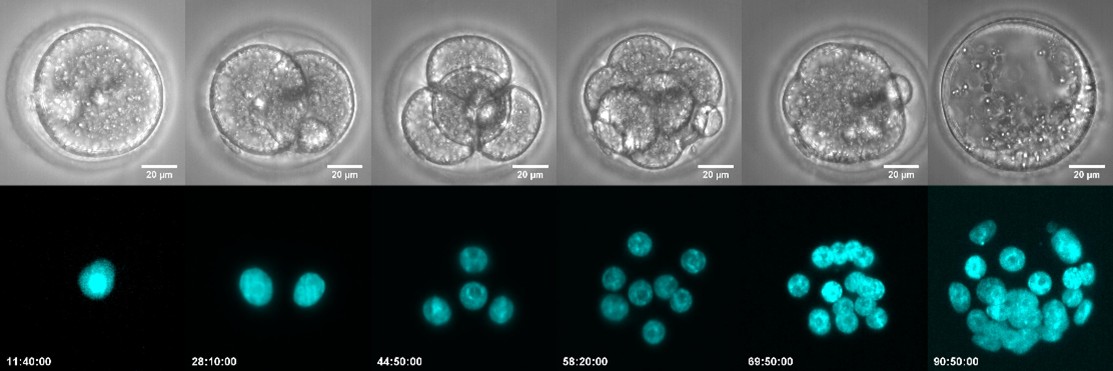
Figure 3: Early development of the mouse embryo from the zygote to the blastocyst stage. The image shows the zygotic stage (one-cell stage), the 2-cell stage, the 4-cell stage, the 8-cell stage, the morula (a compact cluster of cells), and the blastocyst with its blastocoel cavity. Chromatin (DNA) is labeled in green.






